
Problem
Many biologists became a biologist as they love life sciences, and not the computational side of things.
Solution
Having ground up UX development with background in life science domains to develop well-understood UIs for bioinformatics and other life science end users that target the domain expertise of a biologists, not a generic user. Let the scientists do what they do best, and enable them though building the right software.
About Genelinea

Genelinea is an app built at UXBio by biologists, for biologists
When I was back in college, I was employed at the Veterinary Research Institute and worked along side of molecular biologists that were having a hard time with running many of the bioinformatic programs needed to analyze their data. They fully understood what they needed to do, but compiling these foreign programs in Linux distributions with different Perl versions, etc were problematic. I streamlined their workflow by putting together a pipeline of these useful programs. Genelinea focuses on taking sequence data to biological insight, especially when the volume of data overwhelms paradigms for standard data analysis.
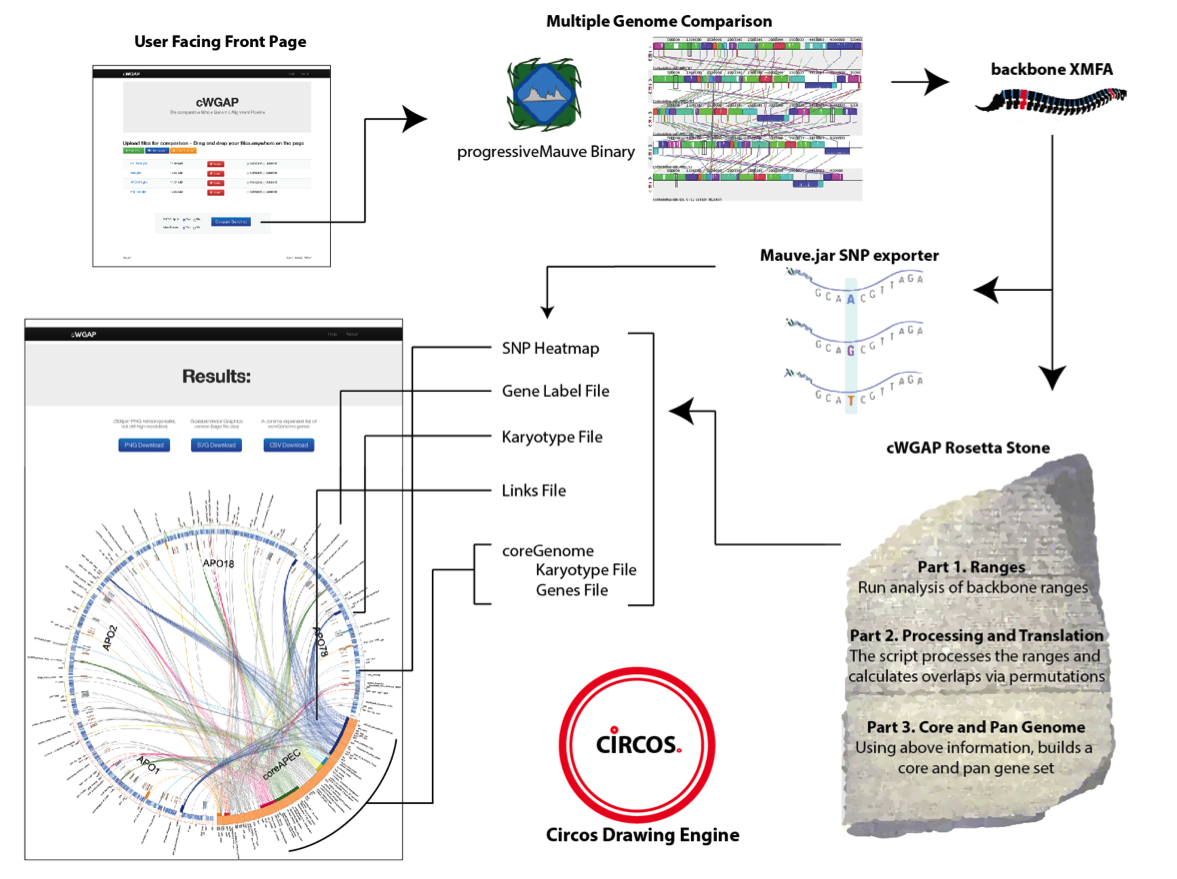
Extending the functionality and visual aspects of comparative genomics programs, while focusing on making the process iterative and easy to use for the biologist enduser. For biologists with limited bioinformatics skills, cWGAP provides an easy to use web-based interface that allows users to upload and compare GenBank files. The backend of cWGAP synchronizes many of the predefined views, options, and paradigms of the datasets entered. cWGAP visually presents the user an ever-growing selection of data output such as core and pan genomic data, SNP data, intergenic link data, and cluster analysis.
How it works
Genelinea is an app built at UXBio by biologists, for biologists
When I was back in college, I worked at the Veternary Research Institute and worked along side of molocular biologists that were having a hard time with running many of the bioinformatic programs needed to analyse their data. They fully understood what they needed to do, but compiling these foreign programs in Linux distribustions with different Perl versions, etc were problematic. I aim to streamline their workflow by putting together a pipleline of these useful programs. Genelinea focuses on taking sequence data to biological insight, especially when the volume of data overwhelms paradigms for standard data analysis.
extending the functionality and visual aspects of comparative genomics programs, while focusing on making the process iterative and easy to use for the biologist enduser. For biologists with limited bioinformatics skills, cWGAP provides an easy to use web-based interface that allows users to upload and compare GenBank files. The backend of cWGAP synchronizes many of the predefined views, options, and paradigms of the datasets entered. cWGAP visually presents the user an ever-growing selection of data output such as core and pan genomic data, SNP data, intergenic link data, and cluster analysis.What it outputs
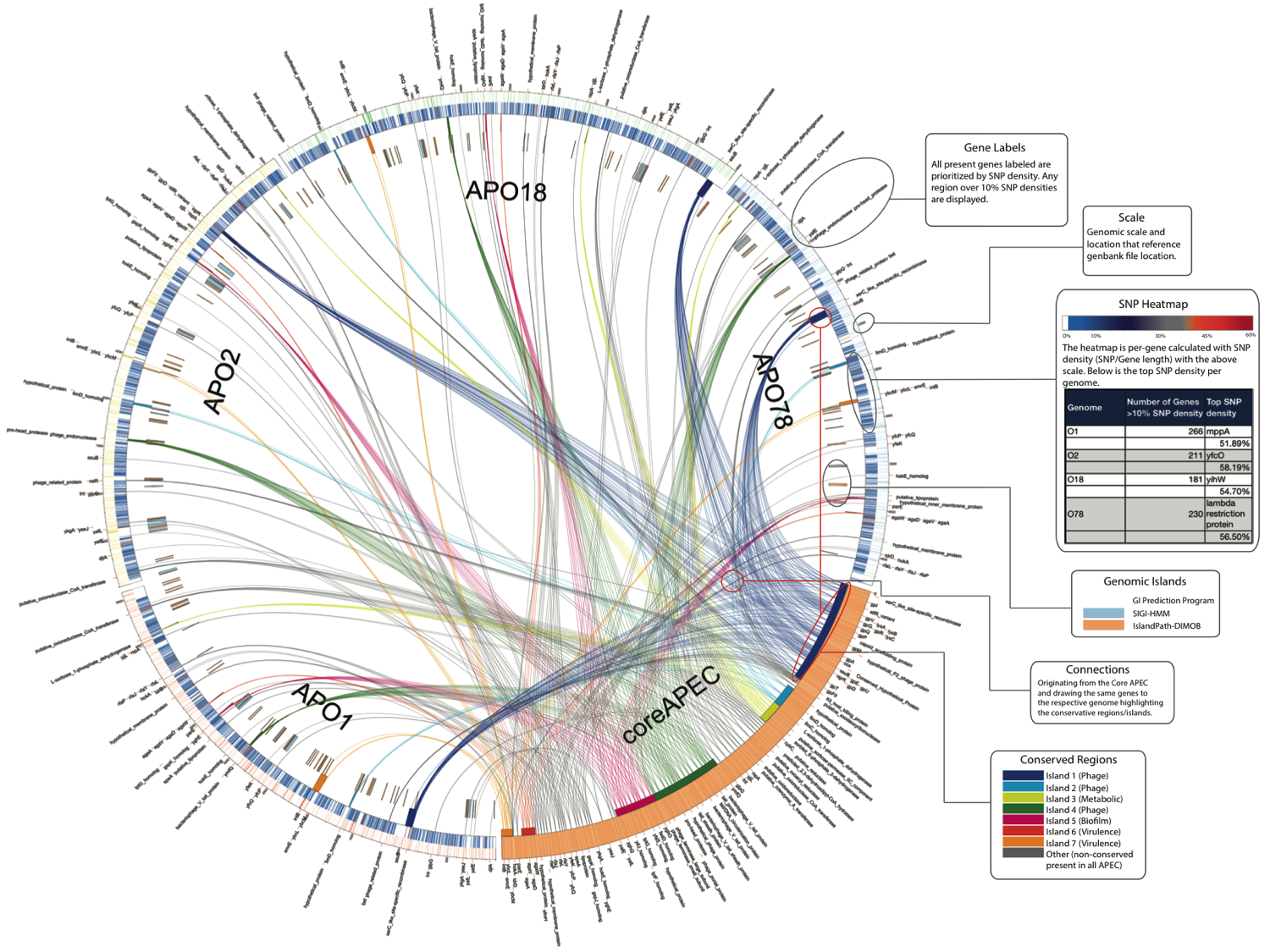
How does it compare to other programs out there?
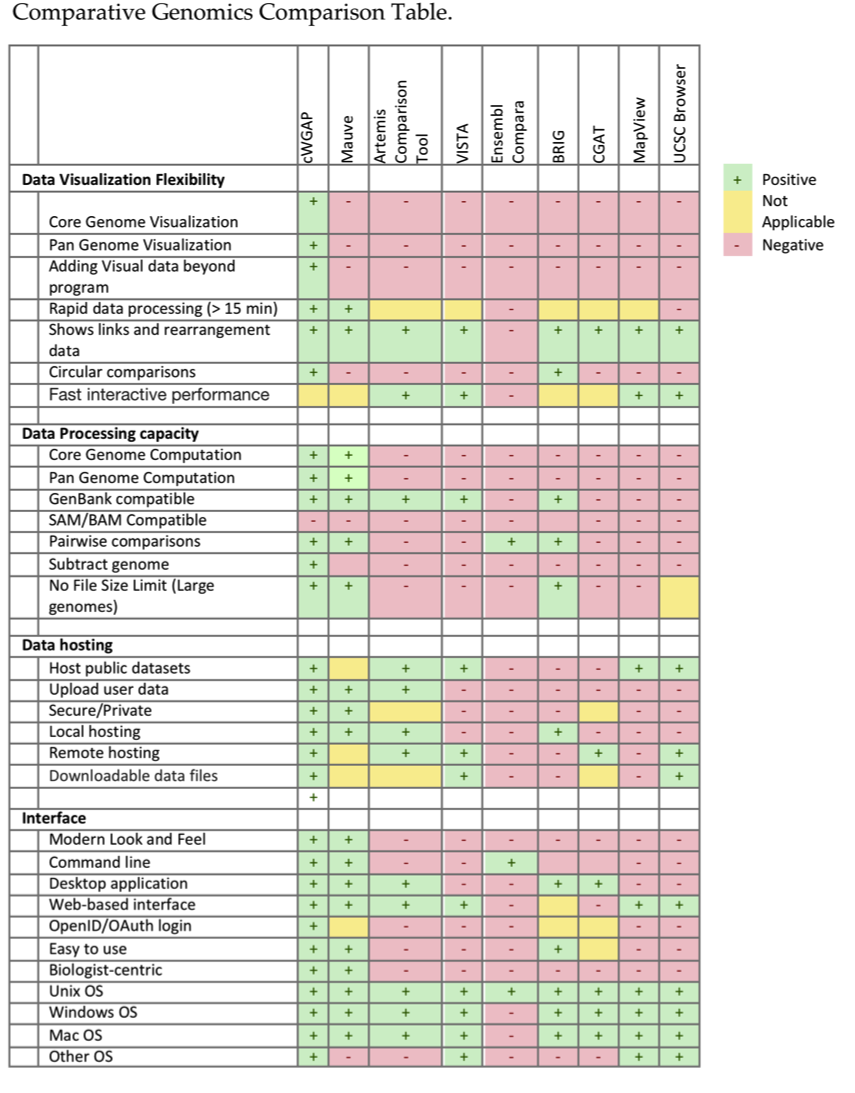
There are not many similar programs, but I put a somewhat overlapping list together
This is not exactly apples to apples, nor apples to oranges. Every workflow may need different end results to display depending on what is being studied. With Genelinea (previously called cWGAP) was created, I tried to make it as broad as possible to answer as many questions as possible for researchers. They have enough decisions and work to do already, I was hoping this would take some time off the overall workflow.
Make an account by logging in with a Google or Twitter account using oauth - or create it the traditional way.

